Walk-Through: Setting up a Student Activity
Let us imagine you want your students to look at the sodium dependence of a standard spike, and you don’t want too many distractions on the way.
Normal extracellular sodium concentration (for a marine squid) is 418 mM. So it seems reasonable to start with a higher than normal concentration (say double, 836 mM), and then successively halve the concentration down to 52 mM, at which point the extracellular sodium concentration will be lower than the intracellular concentration.
The students will measure the peak spike height at each concentration, and plot the results.
- Start Neurosim with the standard HH module (or switch to that module using Model: Hodgkin-Huxley.
Simplify the Setup View
- Select the Options: Configure view menu command to open the Configure dialog.
- Deselect the following check boxes:
Clamp mode choice
Drugs
Patch count
Temperature
Membrane properties
Kinetics.
At this point the dialog looks like this:
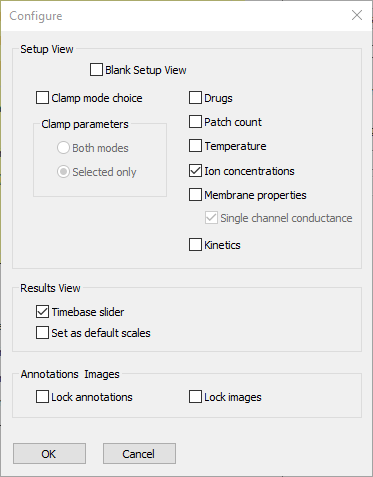
- Click OK to accept the entries and dismiss the dialog.
The Setup view is now much simpler. But be aware that any user could just reverse this process – there is no protection. If you wanted to prevent another user from reversing it, you would use the Lock configuration option available through the Options: Puzzle: Set puzzle menu.
- Increase Pulse 1 Amp to 100.
A bigger stimulus is needed to get a spike at low sodium concentrations. (I don’t know this through some deep wisdom – I know because when I tried this out with the default stimulus, the spike failed.)
- Set Pulse 2 Amp to 0, since no second stimulus is needed.
- Set the extracellular sodium concentration to 836 mM (twice the normal value).
- Click Start to see where we have got to.
Note that the membrane potential (in the upper trace) goes off the screen, so we need to adjust the scales.
- Change the voltage axis upper scale to read 60 and the lower to read -70 (the values are in mV units).
Simplify the Results View
The Results view is showing more traces than are needed for our simple experiment.
- Click Clear to clear the screen. You cannot edit traces with data showing in the Results view.
- Click the Traces button to display the Trace and Axis Setup dialog.
- You cannot edit the Trace setup while the Results view shows data, so click the Clear now button within the dialog. Note that the Clear now button is now hidden. (Alternatively, you could have clicked Clear in the main Results view before clicking Traces).
- Uncheck the Axis box for both Current and Conductance.
- Change the Voltage axis Size parameter to 3.
This will hide both the axes and the traces that normally display on them.
We can leave the colours at their defaults (if we wanted to change one, just click the coloured box and select a new colour).
This axis will display at 3 times the size of any axes with a size of 1, which includes the only remaining visible axis, the Stimulus axis.
At this point the dialog should look like this:
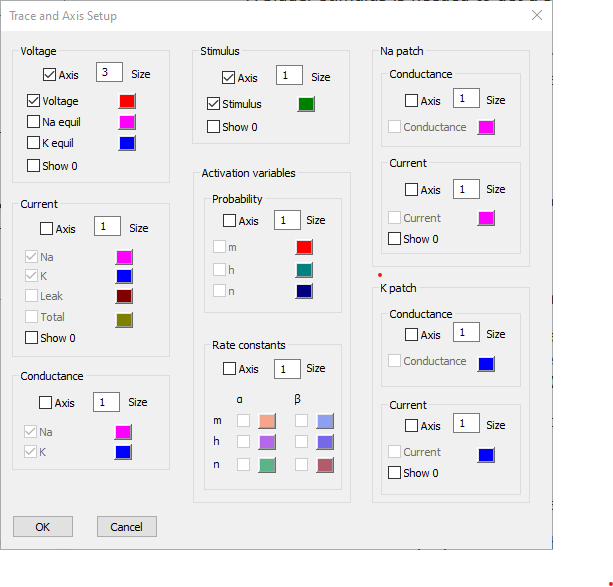
- Click OK.
- Check Run on change box near the top-left of the Results view.
This means that whenever you change an experimental parameter, the simulation will run as though you had clicked Start immediately after making the change. It basically just saves having to repeatedly click Start.
- Select File: Save As (or click the Save As toolbar button
 ) to save these choices to a parameter file that is ready for student use.
) to save these choices to a parameter file that is ready for student use.
For your convenience, a ready-made parameter file sodium dependence is available
The Experiment
Here are instructions for completing the activity.
- Click Start in either the Setup or Results view.
This is necessary even if Run on change is selected, because so far nothing has changed.
A spike should display.
- Change the extracellular sodium concentration to 418 mM (i.e. halve it - this returns it to its normal value).
Run on change was selected in the setup process, so as soon as the user presses Enter or Tab to accept the new concentration value, the simulation should immediately run again. (If Run on change was not selected, just click Start.)
Note that a second, smaller, spike superimposes on the first.
- Repeat the changes in concentration setting the following values:
209
104
52
You should now have 5 spikes of different sizes visible in the Results.
Add Annotations
If the results were being presented in some sort of laboratory report, it would be nice to add some labels to the traces. To do this:
- Right-click somewhere near the top of the biggest spike.
- Select Add annotation from the context menu.
- Enter “836 mM” into the Annotation dialog edit box
The text is a bit small, so
- Click the Font button.
- Set the Size to 10.
- Click OK to dismiss the Font dialog.
- Click OK to dismiss the Annotation dialog.
The text appears in the Results view.
- If the text is not exactly where you want it, drag the annotation text with the mouse to an appropriate location.
- Repeat the process described above to annotate each trace, or:
- Hold down the control key and drag the annotation to a new position.
- Double-click the annotation to access the Annotation dialog,
- or right-click it and select Edit annotation.
- Edit the annotation text appropriately.
This duplicates the annotation.
If you want to adjust the fine position of an annotation
- Click the text to select it. It will be drawn with a box around it.
- Use the keyboard arrow keys to move the text 1 pixel at a time in any direction.
- Click away from the text to de-select it.
The data display in the Results view should now look like this:
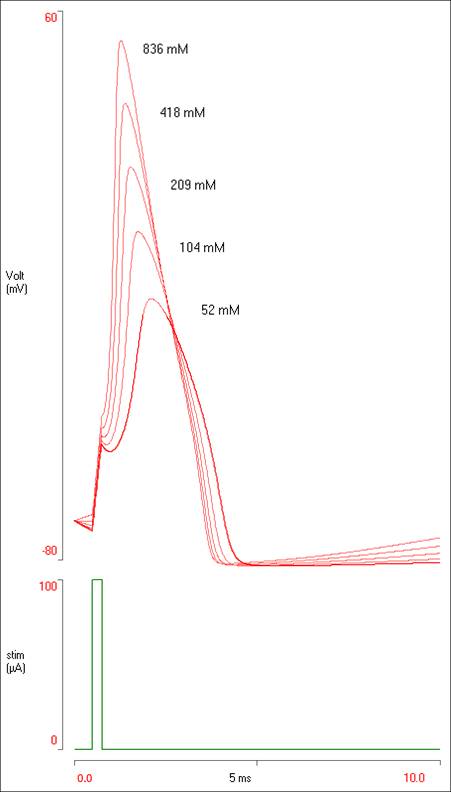
To get a copy of the display like that above:
- Click the Copy button. This copies a bitmap image of the data area onto the clipboard. Then Paste it into your target program.
Note that the Copy button has a drop-down menu associated with it. The upper option, Copy image, does exactly the same as the default action. Copy text places the values of the data themselves onto the clipboard in text format. These could then be pasted into another program for more sophisticated graphing or analysis if desired. Save image and Save text save the image or numerical data to file.
Analyse the Results
- Check the Measure box near the top of the Results view to activate the Measure dialog.
Move the dialog out of the way, so that you can see the Results. - In the Result view, set the Hilight sweep to 1. Now the largest spike is highlighted and the others are greyed out.
- When a sweep is highlighted, measurements are only made from that sweep. If no sweep is highlighted, measurements are made from all sweeps simultaneously. We do not want the latter, because the location of the peak of the action potential varies in each sweep.
- Note: when a sweep is highlighted, the nearby Del[ete] is enabled. Clicking the would delete the highlighted sweep. This is very useful if you have made a mistake in adjusting parameters and want to get rid of just one sweep.
- Click the expand timebase button in the Results toolbar (
 ). This just makes it a bit easier to detect the peak of the spike.
). This just makes it a bit easier to detect the peak of the spike.
HOWEVER, expanding the timebase does not move the annotations, which now probably overlap the traces. This is a useful reminder that annotations are locked to absolute screen positions, not data-relative positions. Restoring the timebase to its original value will put the annotations back where they belong. For now, we must just live with the misplaced annotations. - Drag the red vertical measurement cursor to line up with the peak of the spike. You can use the keyboard arrow keys to fine position it if necessary.
- Click the Measure button in the Measure dialog, and note that the membrane potential value at the spike peak appears in the Voltage column in the dialog.
- Set the Hilight sweep to 2, move the vertical red cursor to the new highlighted spike peak, and repeat the measurement.
- Repeat for each sweep.
At this point you should have 5 rows of measurements in the dialog, with the spike peak at each concentration listed in the voltage column.
- Click +User in the Measure dialog. This inserts a column at the left in which you can enter any data you like.
- Click the left-most column header (the new user column) and enter "[Na]ext" in the Column header dialog. This is not strictly necessary, but it keeps things clearer.
- Click in the first row of the user column, and enter 836.
- Click in the second row (or press the down-arrow on the keyboard) and enter 418.
- Repeat for the remaining rows, entering the appropriate concentration value.
The aim now is to plot the peak of the action potential against the log (base 10) of external the sodium concentration. You could just click Copy in the dialog, and paste the measurements into an external graphing program like Excel. However, we can get a quick preview of the graph in Neurosim itself:
- Click the Plot button in the Measure dialog, to open the XY Scatter dialog.
- In the new dialog, select Voltage from the Y axis drop-down list. You should now see a plot with data points with a declining curve upwards to the right.
- Check the Log10 X box in the Plot dialog.
The data follow a straight line, indicating that the peak spike amplitude is linearly dependent on the logarithm of the sodium concentration gradient, which should be familiar from the Nernst equation. Note that it is linear whether the logarithm is base 10 or base e (the natural log). The value on the X axis changes, but the relationship is linear in either case.
- Uncheck the Connecting line box just below the mid-left of the Plot dialog. The data now display just as a series of dots.
- Check the Linear trendline box near the bottom of the Plot dialog.
A trendline is drawn that closely follows the data. The Plot dialog should now look like this:
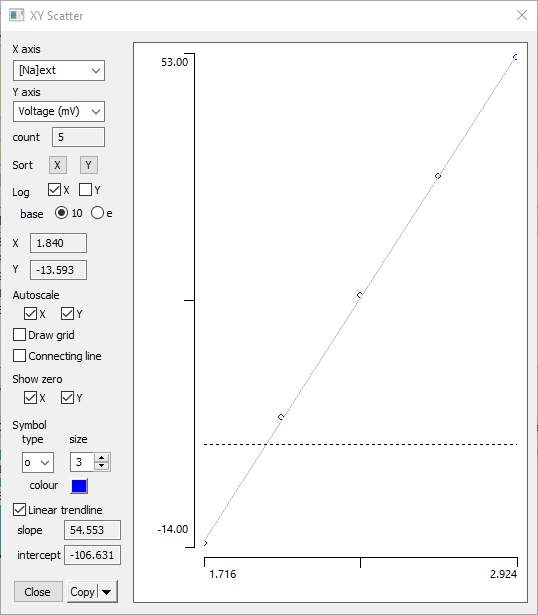
The equation for the trendline is shown just below the Trendline checkbox. Note that the slope is close to 54, which is the Nernst factor at 6°C (the HH experiments were done on a squid from cold sea water: you may be more familiar with 58, which is the factor at room temperature, or 61, which is the factor at mammalian blood temperature).
This tells us that at the peak of the spike the membrane potential is following quite closely to what the Nernst equation would predict for sodium.
- Close the Plot dialog, and then the Measure dialog.
Set a Puzzle
This example is a bit strained, but I want to show the Puzzle facility during the walk-through.
You could imagine asking students to indicate on the figure where they thought the sodium equilibrium potential was relative to the spike. They could not answer this with quantitative accuracy from the information available, but they could certainly give a ball-park indication (somewhat north of the spike peak in each case).
However, they could just ask the program.
- Click Clear.
- Click the Traces button to open the Trace and Axis Setup dialog again.
- Check the Na equil option in the Voltage axis section.
- Click OK.
- Click Start.
The sodium equilibrium potential now shows as a horizontal line just above the peak of the spike. So this answers the question that you want the students to answer.
- Click Clear.
- Select the Options: Puzzle: Set puzzle menu command.
Click OK in the dialog warning about passwords. For this demonstration we don’t need to protect anything.
- In the Traces group within the Puzzle dialog:
- Uncheck the Na equilibrium box.
- Click OK.
- Click Start.
The trace showing the sodium equilibrium potential has disappeared.
If you now access the Trace and Axis Setup dialog by clicking the Traces button as before, you will see (after an alert warning) that the Na equilibrium option is disabled.
If this was being set as a genuine test you wouldn’t want a student to simply reverse the process and re-enable the option. You can prevent this with a password.
- Select the Options: Puzzle: Set password menu command.
- Enter a password (twice).
The user will now not be able to access the Puzzle facility without entering a password first. The password will be stored with any saved parameter file.
WARNING: the password is encrypted and cannot be retrieved if you forget it. You would have to simply re-write the parameter file.
A Final Note
An alert user might notice that at non-standard sodium concentrations, there are changes in membrane potential which start right at the beginning of the simulation, before the stimulus occurs. These occur because there is a non-zero sodium conductance even at rest, so changes in the sodium concentration affect the resting potential, which in turn affect the state of the voltage-dependent channels. It is also worth pointing out that the simulation takes no account of osmotic issues at all. Applying a very high concentration of extracellular sodium might cause a real neuron to just shrivel up and die!
