Bacterial, fungal and viral sialidases

The PDB code links will take you to the PDB entry with links to the publication
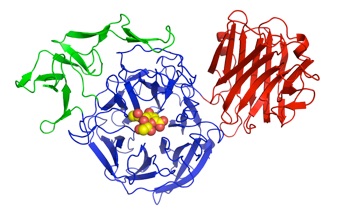
S. pneumoniae encodes three sialdiases, NanA, NanB and NanC. NanA and NanB are present in most strains, and NanA is anchored to the bacterial surface. Below are the structures of teh catalytic domain of NanA, and the complete NanB with its additional carbohydrate-binding domain. This is a collaboration with Peter Andrew, University of Leicester.
Streptococcus pneumoniae
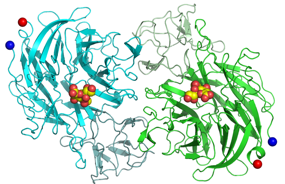
NanA
NanB
Pseudomonas aeruginosa
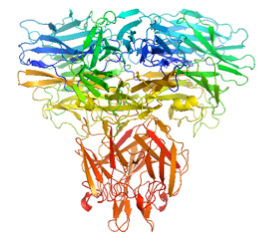
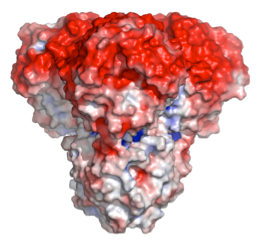
P. aeruginosa encodes a “sialidase”. The structure reveals a trimeric protein held together in part by a TNF-like trimerisation domain. The active site suggests that pseudaminic acid is the substrate, a sialic acid analogue that decorates the pili and LPS of the bacterium.
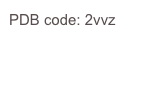
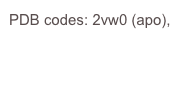
Clostridium perfringens
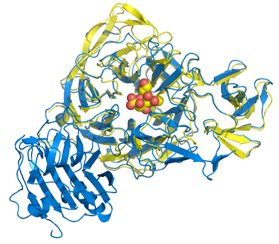
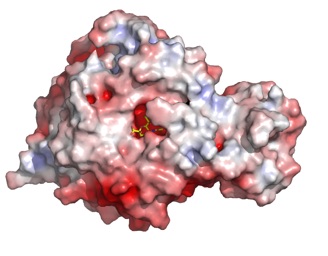
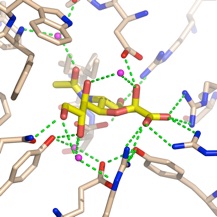
Vibrio cholerae
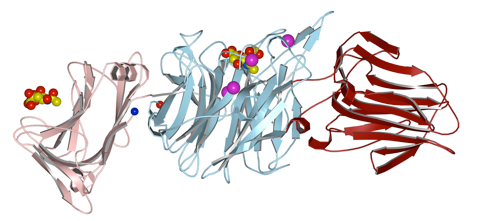
Micromonospora viridifaciens
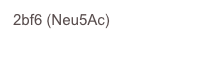


C. perfringens has two sialidases, NanI and NanJ. The crystal structure of the catalytic domain of NanI was determined at atomic resolution in both the apo form and complexed with Neu5Ac. A covalent intermediate was also trapped using a fluorinated sialic acid from Stephen Withers’ group.
V. cholerae sialidase was once known as the ‘receptor destroying enzyme’ given its ability to remove sialic acids from cell surfaces and inhibiting the binding of the influenza virus. It plays a role in pathogenesis in reducing higher order gangliosides to GM1, the receptor for cholera toxin. The structure reveals a catalytic domain flanked by two carbohydrate-binding modules, one of which binds sialic acid.
Sialidases (neuraminidases) remove sialic acid, a 9-carbon carbohydrate that decorates cell surfaces as the terminal sugar of various glycoconjugates. Baterial sialidases come in may sizes. Some just contain the beta-propeller catalytic domain, others have additional carbohydrate-binding modules (CBMs) that often recognize sialic acid and help the enzyme target its substrate on cell surfaces. Most are monomers, but below we have examples of a dimer and a trimer. We are engaged in structure-based drug discovery of the S. pneumoniae and P. aeruginosa enzymes, knockouts of which severely reduce pathogenicity in mouse models. NMR studies have been carried out in collaboration with Jenny Wilson & Milton Kiefel, Griffith University, Australia.
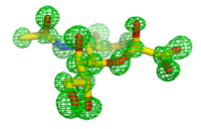

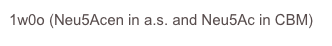

This soil bacterium secretes two forms of sialidase from the same gene. A short form, with just the catalytic domain, and a longer form with a galactose-binding module, dependent on the food source. The galactose-binding domains is linked via an immunoglobulin-like domain.
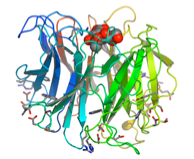



As series of mutants of the larger form (68kDa) were studied in collaboration with Andy Bennet, Simon Fraser University, BC, Canada, to probe the function of catalytic residues.



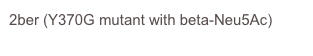

Salmonella typhimurium
A small 42kDa sialidase that was the the first reported structure of a bacterial sialidase showing the similarity to the influenza virus neuraminidase.
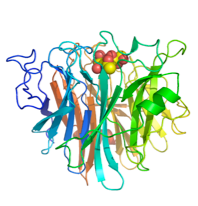




Aspergilus fumigatus
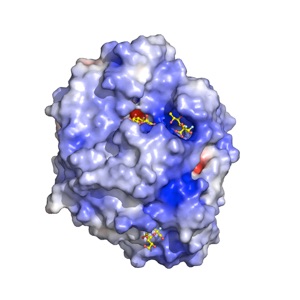
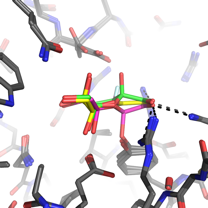
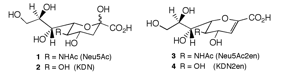
We recently discovered that the A. fumigatus sialidase is a KDNase, and possibly recognises polyKDN. KDN is Neu5Ac with the acetyl group replaced by a hydroxyl. The crystal structures of the enyzme in complex with KDN, an inhibitor and a flourinated substrate have enables capture of the catalytic cycle. This is a collaboration with Margo Moore and Andy Bennet at Simon Fraser, BC, Canada.