Cleavage Activity of Aphtho- and Cardiovirus 2A Oligopeptidic Sequences.Staff Involved;
Dr. Andriy Sukhodub (Post-Doctoral Research Fellow) - cellular 2A-like sequences
Dr. Garry Luke (Post-Doctoral Research Fellow) - cellular 2A-like sequences
Dr. Ekaterina Miskaia (Post-Doctoral Research Fellow) - 2A in human gene therapy applications
Ms. Valerie Odon (PhD student) - mechanism of 2A and 2A-like sequences
Mr. John Nicholson (Technician).
In the entero- and rhinoviruses the 1D/2A primary cleavage is mediated by a well characterised virus encoded proteinase (2Apro), of some 17kDa, acting in an intramolecular fashion (in cis) to cleave the nascent polyprotein at its own N-terminus.
The analogous primary cleavage in many other genera of the Picornaviridae (aphtho-, cardio-, erbo-, tescho-, cosa- and some parechoviruses) occurs at the C-terminus of the 2A region between the capsid protein precursor ([P1-2A] - aphthoviruses; [L-P1-2A] - cardioviruses) and 2BC / P3.
Inspection of the cardiovirus 2A protein sequence (ca 15kDa) reveals no similarity to 2Apro of the entero- and rhinoviruses and none of the characteristic proteinase sequence motifs. The 2A region of the aphthovirus foot-and-mouth disease virus (FMDV) is only 18aa long - but is highly similar to the C-terminal region of cardiovirus 2A.
Our previous work on the function of the 2A region has demonstrated a number of important features;
(i) The FMDV 2A sequence (together with the N-terminal proline of protein 2B) retained ‘cleavage’ activity in recombinant FMDV polyproteins when either the upstream or downstream contexts were replaced, but the upstream context influenced the activity (Ryan et al., 1991).(ii) A single ORF encoding an artificial polyprotein comprising FMDV 2A (plus the N-terminal proline of protein 2B; 19aa in total) flanked by the reporter proteins CAT (chloramphenicol acetyl-transferase) and GUS (beta-glucuronidase) was constructed.


These cDNA constructs were analysed using in vitro translation systems. The translation profile obtained from pCATGUS showed the expected uncleaved product whereas the translation profile obtained from pCAT2AGUS showed three major translation products - uncleaved [CAT2AGUS], GUS and [CAT2A]. The FMDV 2A sequence was, therefore, able to mediate a co-translational ‘cleavage’ in this artificial polyprotein directly analogous to its function in FMDV polyprotein processing: ~95% of the radiolabeled translation product was in the ‘cleaved’ form (Ryan and Drew, 1994).
(iii) 2A-mediated ‘cleavage’ occurred only co-translationally - upon prolonged incubation the ‘uncleaved’ translation products did not subsequently cleave (Ryan & Drew, 1994).(iv) The C-terminal region (19aa) of the 2A protein of the cardioviruses encephalomyocarditis virus (EMCV) and Theiler's Murine Encephalopathy virus (TME), together with the completely conserved N-terminal proline of protein 2B, was as active as the FMDV 2A sequence (Donnelly et al., 1997).
(v) The artificial [CAT2AGUS] polyprotein did not ‘cleave’ when expressed in prokaryotic systems (Donnelly et al., 1997).
Our initial working hypothesis was that this short 2A region could mediate a single-turnover proteolysis of the polyprotein, in cis, at the 2A/2B site (invariantly a glycine / proline pair). The 2A sequence, together with the N-terminal residue of 2B, would represent an autonomous, self-aligning, nucleophile:electrophile couple that brought about the cleavage of the Gly-Pro peptide bond (not a proteinase:substrate couple sensu stricto).
In this hypothetical senario the sequences in a proteinase which are concerned with imparting substrate specificity could be dispensed with. Similarly, the sequences required to provide the molecular environment whereby the active site nucleophile may be regenerated could also be dispensed with. Thus one might envisage such a short sequence could bring about this specific proteolytic event.
Careful analysis of the translation profiles of the [CAT2AGUS] self-processing artificial polyprotein system showed considerable internal initiation within the CAT sequence in coupled transcription / translation in vitro systems (Donnelly et al., 1997).
The co-translational ‘cleavage’ of the [CAT2AGUS] polyprotein into the [CAT2A] and GUS products was monitored by phosphorimaging. In both rabbit reticulocyte lysate and wheat germ extract in vitro translation systems an imbalance in the accumulated translation products was observed.
Our hypothesis that 2A functions as a proteolytic element, however, would predict a 1:1 stoichiometry of the cleavage products.We have performed detailed analyses of the translation profiles from three types of polyprotein. The first is artificial self-processing polyproteins comprised of two reporter proteins flanking FMDV 2A. The second, an FMDV polyprotein in which 2A is in its native context. Lastly, a polyprotein containing a defined cis-acting proteinase derived from human rhinovirus.
We have shown striking differences in the polyprotein processing properties of these systems. Whilst the proteolytically processed human rhinovirus polyprotein showed equimolar quantities of the cleavage products, the artificial polyproteins showed a molar excess of the translation product N-terminal of the 2A sequence to that C-terminal of 2A.
We have performed experiments designed to investigate the reason for this imbalance:
(i) protein degradation studies have shown that neither CAT nor GUS are degraded at levels anywhere nearly enough to explain our imbalance,
(ii) reversing the gene order within the artiificial polyprotein (GUS2AGFP) results in a molar excess of GUS over GFP,
(iii) the translational or transcriptional properties of the coupled (transcription/translation) in vitro systems were investigated. Translation of the P1-P2 region of human rhinovirus 14, cleaved at the P1/P2 junction by the 2A proteinase, showed the endogenous HRV P1/P2 processing does yeild a 1:1 ratio of cleavage products. Note: the HRV P1/P2 ORF is some 50% longer than the GFP2AGUS artificial polyproteins described above. These data indicate that is possible to synthesise long open reading frames without appreciable non-specific termination of either transcription or translation.
A model of the FMDV 2A ‘cleavage’ was developed in which the oligopeptide sequence is proposed to act as a cis-acting hydrolyase element (CHYSEL), although not as a novel cis-acting proteolytic element, but as an esterase (Ryan et al., 1999; Donelly et al., 2001a,b).
Translational Model of 2A Activity.
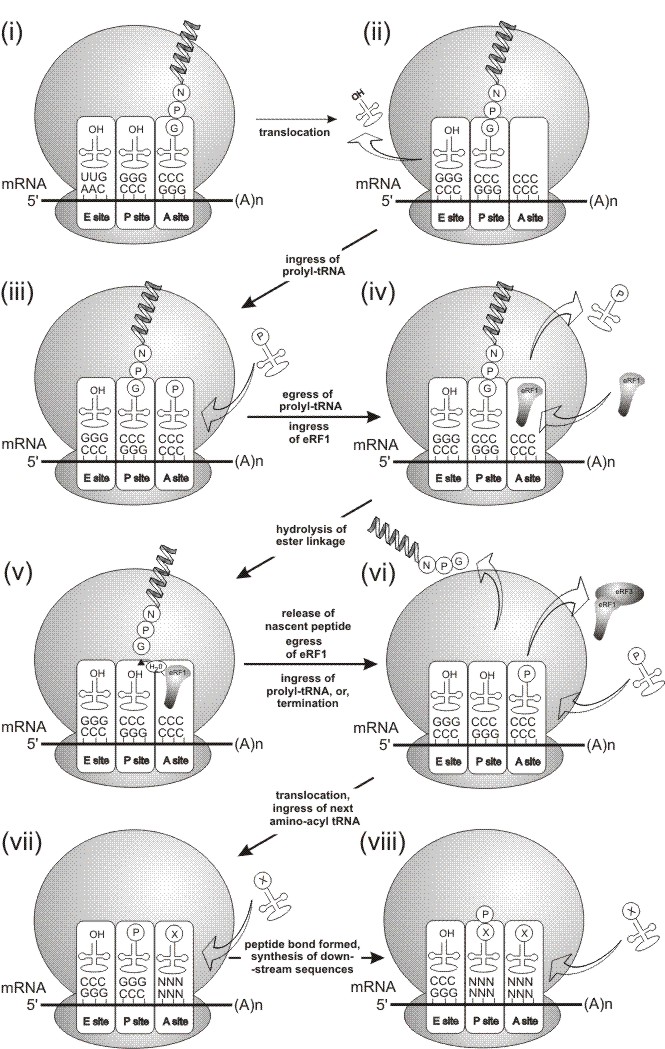
Mechanism of 'Cleavage'. The mechanism that lead to the (proposed) hydrolysis of the peptidyl:glycyl-tRNA ester linkage was not understood.
Recently, however, the involvement of transtion release (termination) factors 1 and 3 in the 'cleavage' mechanism was demonstrated (Doronina et al., 2008a,b.). These proteins bind to stop codons and are resposible for the hydrolysis of the ester linkage between the ultimate amino acid in the nascent peptide chain and it's tRNA. In the case of 2A, the translation complex is arrested at the C-terminus of 2A (within the ORF).
The 2A nascent peptide and glycyl-tRNA is translocated from the A to P site (steps i & ii, below).
Prolyl-tRNA enters the A site (step iii), but we propose that the prolyl-tRNA in the A site is unable to form the peptide bond and exits the A site (step iv).
eRF1 enters the A site and hydrolyses the ester linkage, releasing the nascent peptide (steps iv and v).
Following the hydrolysis of the ester linkage, the nascent peptide is released. eRF1 leaves the complex, eRF3 being involved in this process (steps v and vi).
Two, mutually exclusive, outcomes may then arise. One outcome is that translation terminates at the C-terminus of 2A. The alternative outcome is that eRF1 exits the A site, proly-tRNA (re-)enters the A site, is translocated by eEF2 into the P site and the next amino-acyl tRNA can enter the A site to synthsise the downstream sequences (steps vi to viii).
Data from the Atkins and Doronina papers show the N-terminal residue of the protein downstream of 2A is proline. Data from the Doronina paper shows the C-terminal residue of 2A is glycine: all amino acids encoded by the mRNA may be synthsised - it is just the synthesis of the glycyl-prolyl peptide bond that may be 'skipped'. Ribosome toe-printing data from 2A-paused translation complexes maps the site of pausing to exactly that predicted by the model proposed in Donnelly et al., (2001a).
This translational model accounts for the excess of translation product upstream of 2A over that downstream of 2A we observe using in vitro translation systems.
Expression of 2A-containing artificial polyproteins in vivo shows, however, that the translation products are synthesised in approximately equal amounts.
2A-Like Sequences
We have identified a number of '2A-like' sequences from other viruses; a range of positive-stranded RNA insect viruses, dsRNA insect viruses, a dsRNA crustacean virus, dsRNA mammalian viruses (type C rotaviruses) and repeated sequences (L1 non-LTR retrotranspsons) within Trypanosoma spp. All of these 2As have been tested and found to mediate cleavage (Donnelly et al., 1997; Donnelly et al., 2001b; Heras et al., 2006; Luke et al., 2008).
A small number of '2A-like' have been identified in cellular genes, although none of these show any 'cleavage' activity (Donnelly et al., 2001b).
Publications;
Doronina, V.A., Wu, C., de Felipe, P., Sachs, M.S., Ryan, M.D. & Brown, J. (2008a). Site-specific release of nascent chains from ribosomes at a sense codon. Mol. Cell. Biol. 28, 4227- 4239.
Doronina, V.A., de Felipe, P., Wu, C., Sharma, P., Sachs, M.S., Ryan, M.D. & Brown, J.D. (2008b). Dissection of a co-translational nascent chain separation event. Biochem. Soc. Trans. 36, 712-716.
Luke, G.A., de Felipe, P., Lukashev, A., Kallioinen, S.E., Bruno, E.A & Ryan, M.D. (2008). The occurrence, function, and evolutionary origins of ‘2A-like’ sequences in virus genomes. J. Gen. Virol. 89, 1036-1042.
Atkins, J.F., Wills, N.M., Loughran, G., Wu, C-Y., Parsawar, K. Ryan, M.D., Wang, C.H. & Nelson, C.C. (2007). A case for "StopGo": Reprogramming translation to augment codon meaning of GGN by promoting unconventional termination (Stop) after addition of glycine and then allowing continued translation (Go). RNA 13, 1-8.
Heras, S.R., Thomas, M.C., García, M., de Felipe, P. García-Pérez, J.L., Ryan, M.D & López, M.C. (2006). L1Tc non-LTR retrotransposons from Trypanosoma cruzi contain a functional viral-like self-cleaving 2A sequence in frame with the active proteins they encode. Cell Mol Life Sci. 63, 1449-1460.
de Felipe, P., Luke, G.A., Hughes, L.E., Gani, D., Halpin, C. & Ryan, M.D. (2006). E unum pluribus: multiple proteins from a self-processing polyprotein. Trends in Biotechnology 5, 68-75.
Osborn, M.J., Panoskaltsis-Mortari, A., McElmurry, R.T., Bell, S.K., Vignali, D.A.A., Ryan, M.D., Wilber, A., McIvor, R.S., Tolar, J. & Blazar, B.R. (2005). A picornaviral ‘2A-like’ sequence based tricistronic vector allowing for high level therapeutic gene expression coupled to a dual reporter system. Mol. Therapy 12, 569-574.
de Felipe, P & Ryan, M.D. (2004) Targeting of proteins derived from self-processing polyproteins containing multiple signal sequences. Traffic 5, 616-626.
El Amrani, A., Barakate, A., Askari, B.M., Li, X., Roberts, A.G., Ryan, M.D. & Halpin, C. (2004) Coordinate expression and independent subcellular targeting of multiple proteins from a single transgene. Plant Physiol. 135, 16-24.
de Felipe, P. Hughes, L.E. Ryan, M.D. & Brown, J.D. (2003). Co-translational, intraribosomal cleavage of polypeptides by the foot-and-mouth disease virus 2A peptide. J. Biol. Chem. 278, 11441-11448.
Halpin, C., Barakate, A., Askari, B., Abbott, J. & Ryan, M.D. (2001). Enabling technologies for manipulating multiple genes on complex pathways. Plant Mol. Biol. 47, 295-310.
Klump, H., Schiedlmeier, B., Vogt, B., Ryan, M.D., Ostertag, W. & Baum, C. (2001). Retroviral vector-mediated expression of HoxB4 in hematopoietic cells using a novel coexpression strategy. Gene Therapy 8, 811-817.
Donnelly, M.L.L., Luke, G., Mehrotra, A., Li, X., Hughes, L.E., Gani, D. & Ryan, M.D. (2001a). Analysis of the aphthovirus 2A/2B polyprotein 'cleavage' mechanism indicates not a proteolytic reaction, but a novel translational effect: a putative ribosomal 'skip'. J. Gen. Virol. 82, 1013-1025. 30. (download this paper)
Donnelly, M.L.L., Hughes, L.E., Luke, G., Li, X., Mendoza, H., ten Dam, E., Gani, D. & Ryan, M.D. (2001b). The 'cleavage' activities of FMDV 2A site-directed mutants and naturally-occurring '2A-Like' sequences. J. Gen. Virol.82, 1027-1041. (download this paper)
Luke, G & Ryan M.D. (2001). Translating the message. Biologist 48, 79-82.
Halpin, C., Barakate, A., Askari, B., Abbott, J. & Ryan, M.D. (2001). Enabling technologies for manipulating multiple genes on complex pathways. Plant Mol. Biol.47, 295-310.
Klump, H., Gruettner, C., Rimek, A., Ryan, M.D., Ostertag, W. & Baum, C. (2001). The 2A proteinase of Foot-and-Mouth Disease Virus (FMDV) is superior to Internal Ribosomal Entry Sites (IRESes) for co-expression strategies using retroviral vectors. Gene Therapy 8, 811-817.
Ryan, M.D., Donnelly, M.L.L., Lewis, A., Mehrotra, A.P., Wilkie, J. & Gani, D. (1999). A model for non-stoichiometric, co-translational protein scission in eukaryotic ribosomes. Bioorganic Chem. 27, 55-79.
Ryan, M.D., Donnelly, M.L.L. & Gani, D. (1998). Aphtho-, cardiovirus 2A autolytic sequence. In ‘A Handbook of Proteolytic Enzymes’. Barrett, A. and Rawlings, N.J. eds. pp 1598-1600. Academic Press, London.
Donnelly, M., Gani, D., Flint, M., Monoghan, S. & Ryan, M.D. (1997). The cleavage activity of aphtho- and cardiovirus 2A proteins. J. Gen. Virol.78, 13-21. (Download this paper)
Hartzoulakis, B., Rutherford, T.J., Ryan, M.D. & Gani, D. (1996). Synthesis and properties of a biocompatible analogue for b-turn protein structural motifs based on 5-amino-3-pentynoic acid. Tetrahedron Letters 37, 6911-66914.
Ryan, M.D. & Drew, J. (1994). Foot-and-mouth disease virus 2A oligopeptide mediated cleavage of an artificial polyprotein. EMBO J. 134, 928-933.
Ryan, M.D., King, A.M.Q. & Thomas, G.P. (1991). Cleavage of foot-and-mouth disease virus polyprotein is mediated by residues located within a 19 amino acid sequence. J. Gen. Virol.72, 2727-2732.